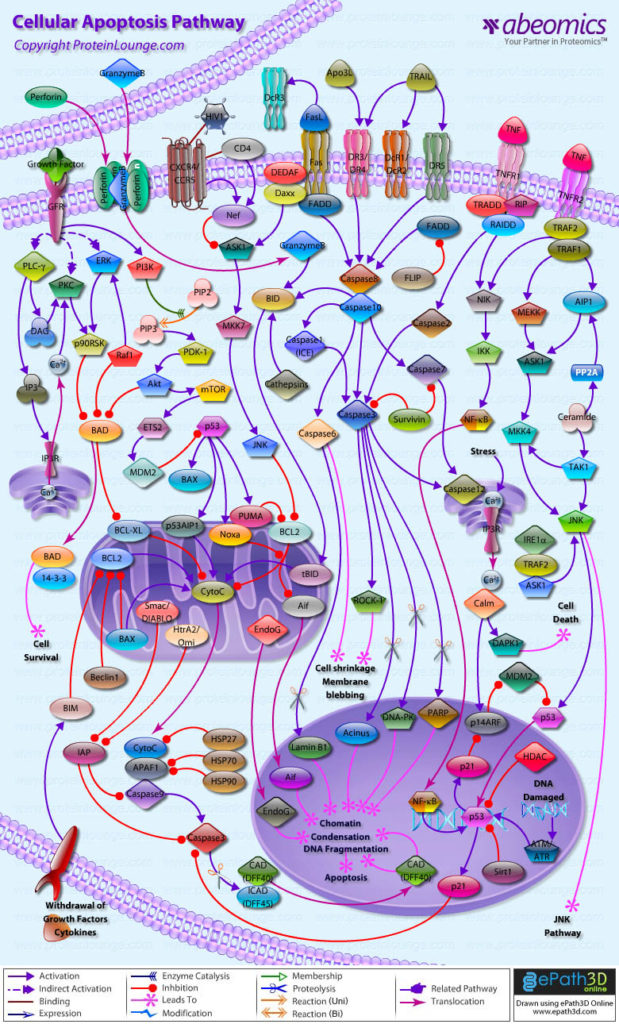
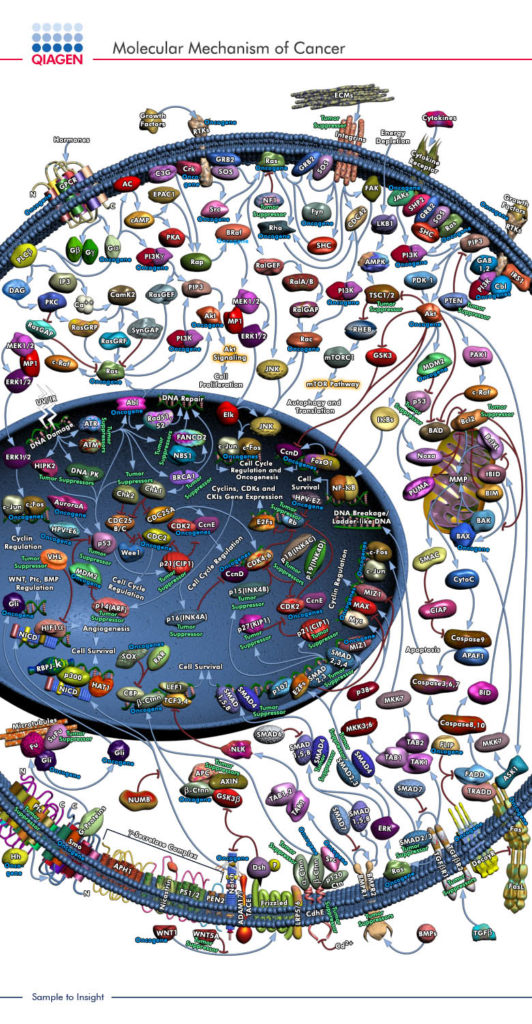
In molecular and cellular biology there are two three dimensional aspects that are often portrayed in two dimensions, these are molecules and their interaction within pathways. Molecules are usually drawn as simple blobs or shapes in order to better visualize the different domains that have specific functions (proteins have very complex shapes). With these simple shapes, their interactions are usually connected with a vast map of arrows and inhibitions which form the pathways that biologists study and use to develop function specific drugs or site specific research. This decision to go simple and 2D has made studying and using biology a lot simpler but it has taken out a very important factor, molecules and pathways have movement. This movement, which is important in the interaction of proteins, depends vastly on size and surroundings which are all three dimensional. Naturally, to model this mathematically is quite complex and requires a lot of prior knowledge and computational power but once achieved has great benefits. If these pathways could be brought into a dynamically moving intractable three dimensional world, then there would be the possibility of better research and understanding into medicine. By better understanding what happens when certain interactions are “physically” and “visually” removed there would be less wasted effort in pathway structure and experimental design.
Check out this cancer pathway map: https://www.qiagen.com/dk/shop/genes-and-pathways/pathway-details/?pwid=301